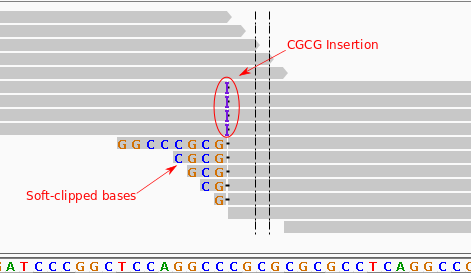
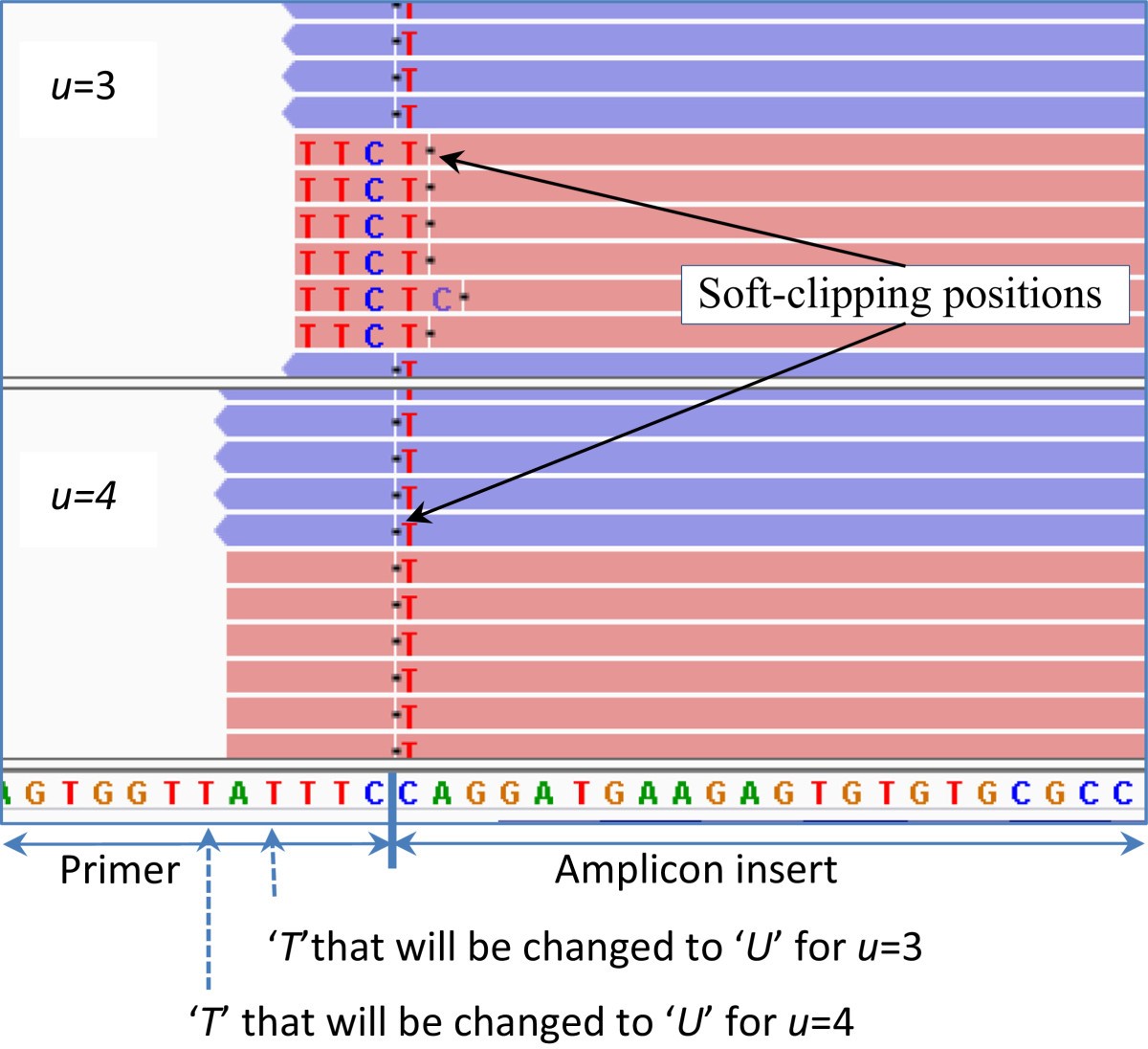
Soft-clipped reads vs. improperly aligned reads at a structural variant... | Download Scientific Diagram

IMSindel: An accurate intermediate-size indel detection tool incorporating de novo assembly and gapped global-local alignment with split read analysis | Scientific Reports
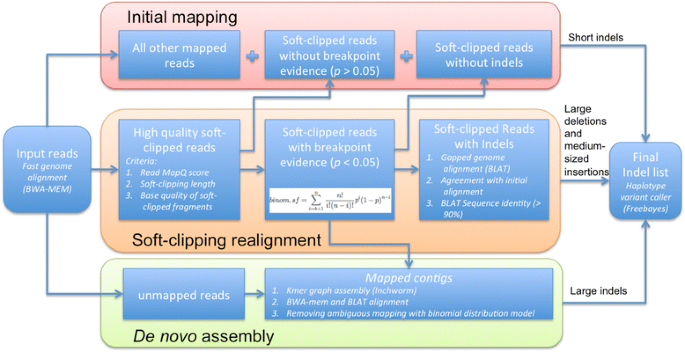
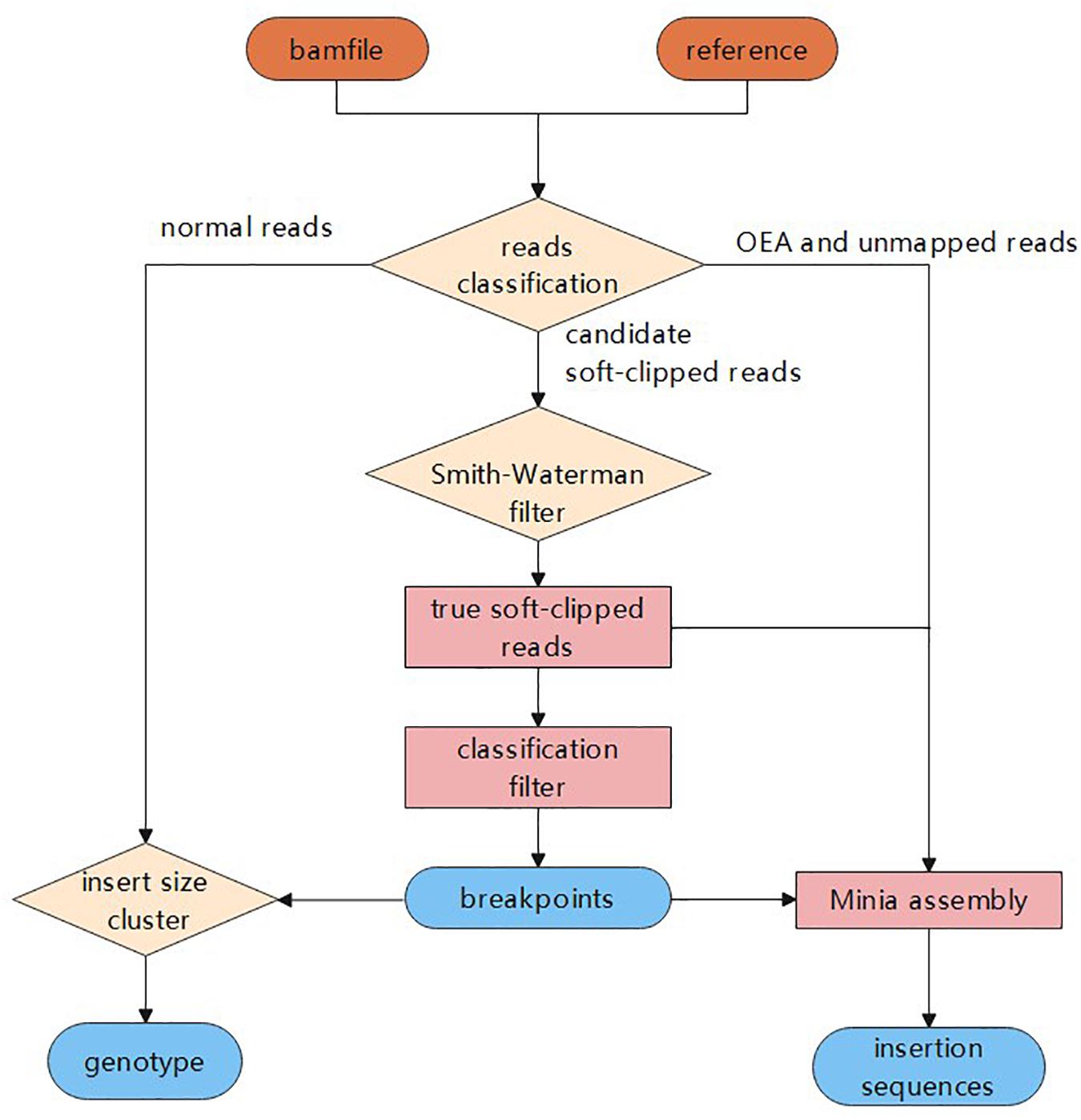
ScanIndel: a hybrid framework for indel detection via gapped alignment, split reads and de novo assembly | Genome Medicine | Full Text
SoftSearch: Integration of Multiple Sequence Features to Identify Breakpoints of Structural Variations | PLOS ONE
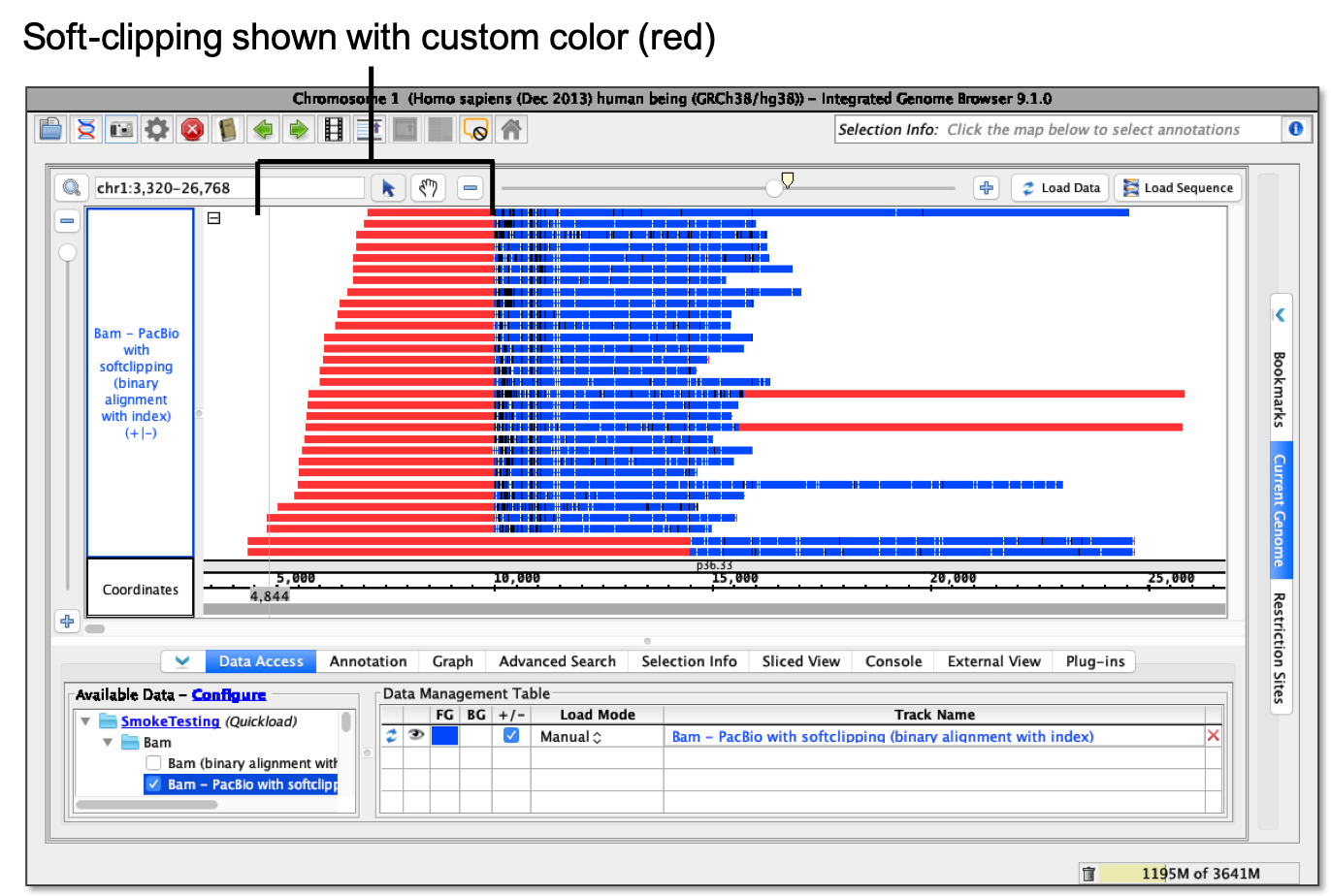
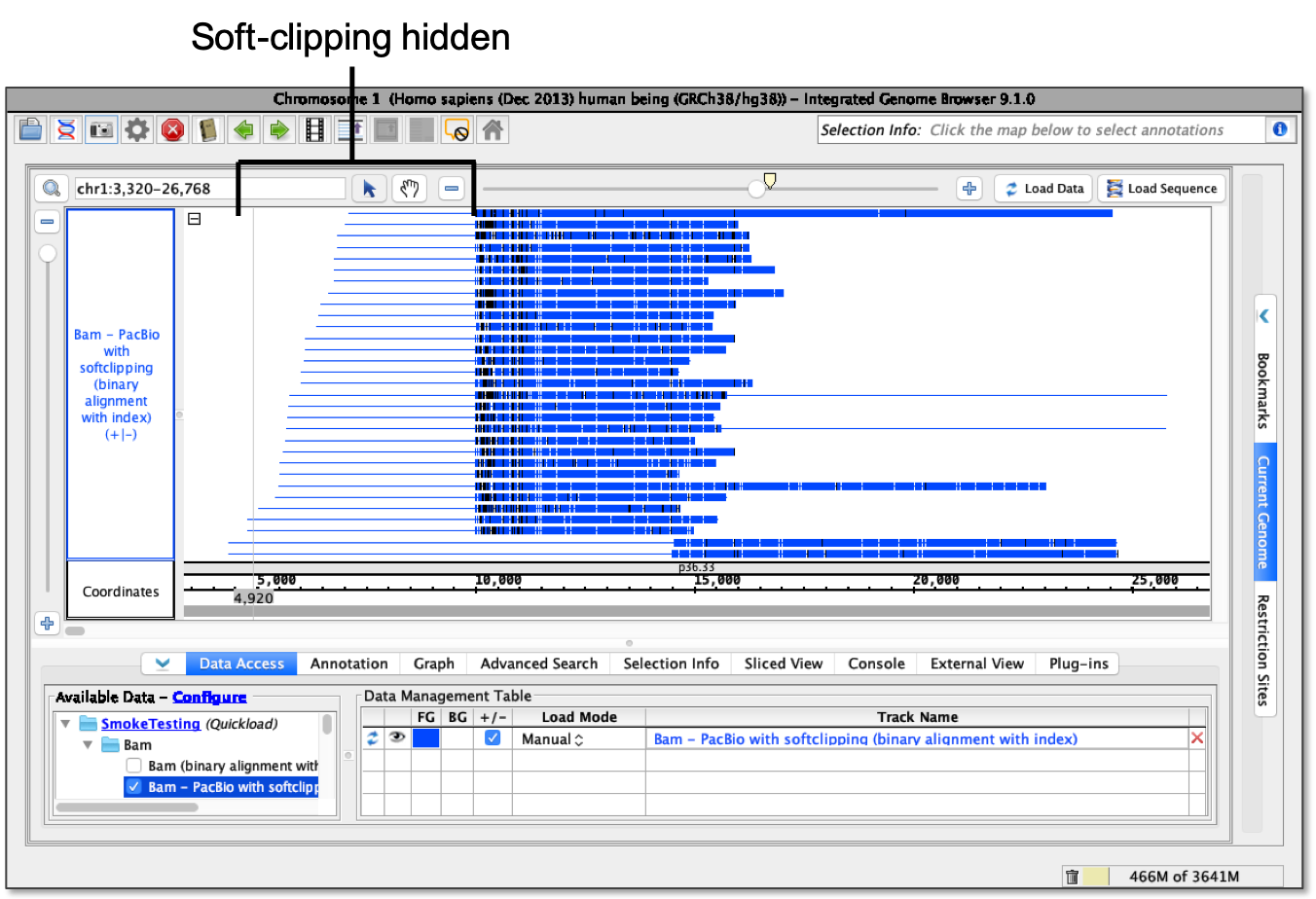
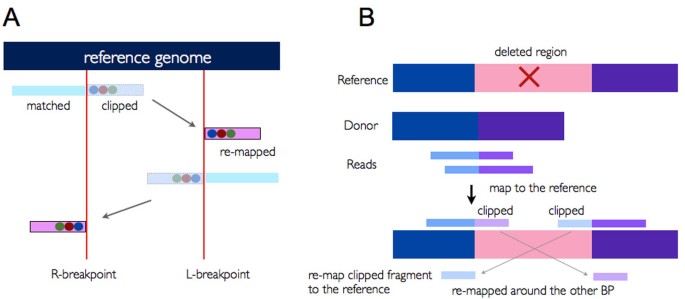
QC Fail Sequencing » Soft-clipping of reads may add potentially unwanted alignments to repetitive regions
ClipCrop: a tool for detecting structural variations with single-base resolution using soft-clipping information | BMC Bioinformatics | Full Text
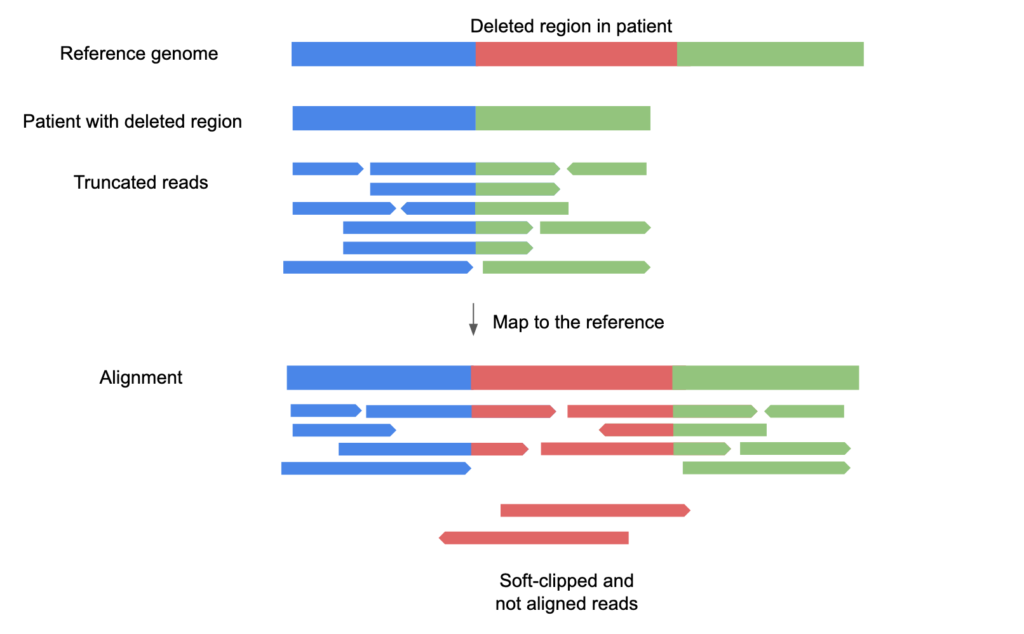
Challenges and importance of mid-sized deletion identification for genomic medicine - SeqOne Genomics

Soft-clipped reads vs. improperly aligned reads at a structural variant... | Download Scientific Diagram

Soft-clipped reads vs. improperly aligned reads at a structural variant... | Download Scientific Diagram
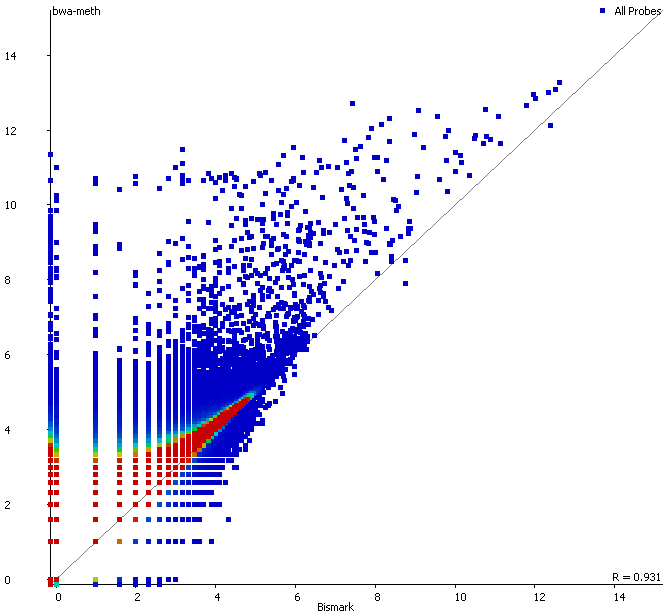
QC Fail Sequencing » Soft-clipping of reads may add potentially unwanted alignments to repetitive regions
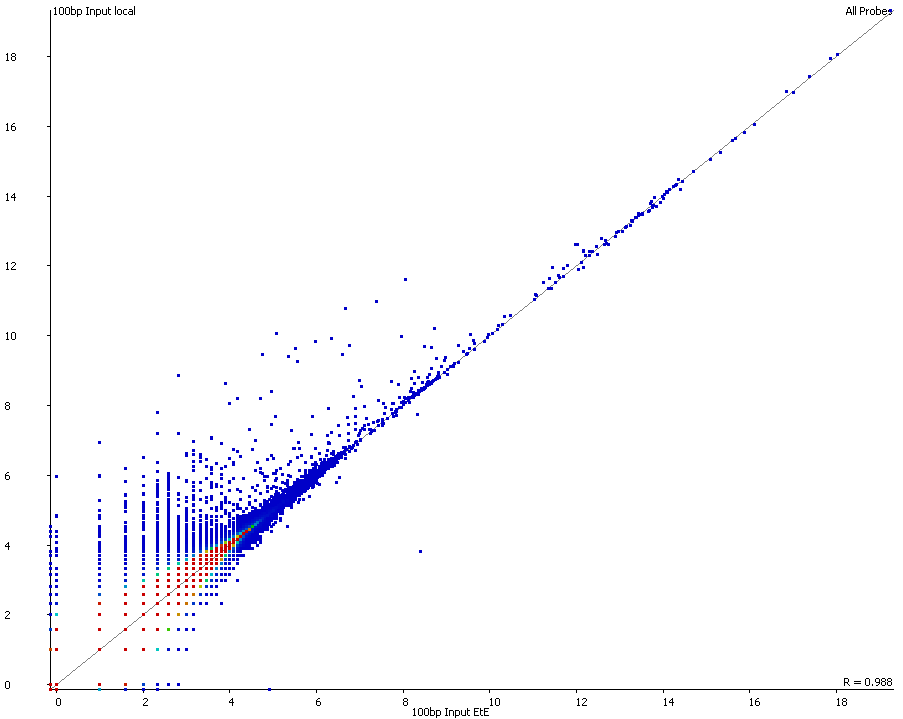
QC Fail Sequencing » Soft-clipping of reads may add potentially unwanted alignments to repetitive regions

Soft-clipped reads vs. improperly aligned reads at a structural variant... | Download Scientific Diagram